A new method to describe electron density based on machine learning
This last month an article from our group titled “Analytical Model of Electron Density and Its Machine Learning Inference” was accepted in the Journal of Chemical Information of Modeling.
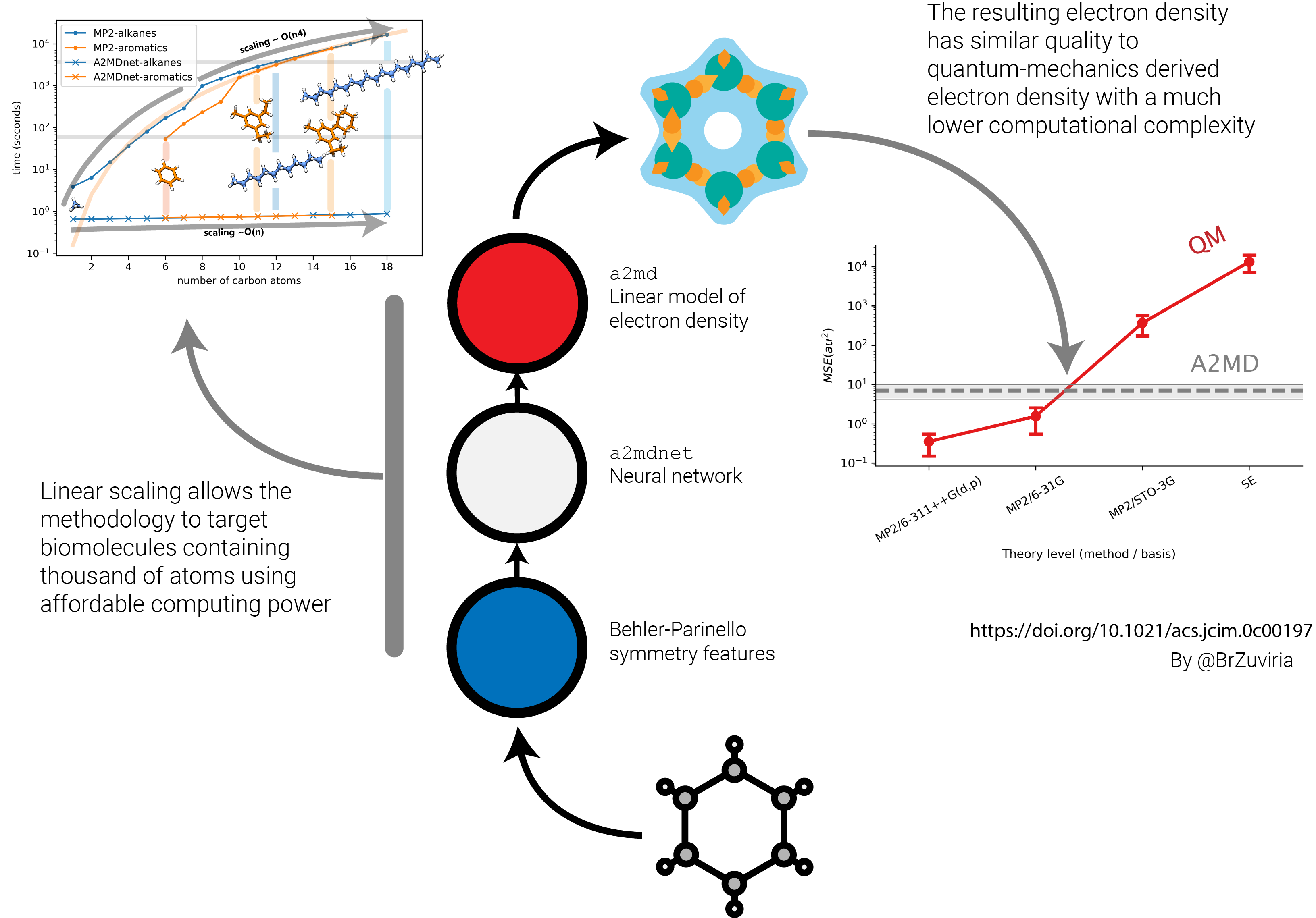
This article belongs to our new line of research of methods for theoretical chemistry. It explains a methodology born by combining linear models of electron density with machine learning to provide, among other advantages, a technique with a quasi-linear scaling. This can help to describe the electron density of biomolecules containing thousands of atoms without requiring much computational power. The electron density is the probability of finding an electron in a given region of space. It is the squared product of the electron wavefunction, and, according to Kohn theorems, it provides the information from which to derive all the molecular properties of a molecule in its ground state.
In our methodology, we provide a linear model of electron density based on a sum of exponentials combined with anisotropic angular terms to describe bonding-electron density deformation. Then, a neural network was trained to reproduce the parameters of each molecule. The result is a light model of electron density which can be fastly generated without using more than its molecular graph and coordinates.
The code is publicly available at https://github.com/brunocuevas/a2md